| Use the HUF-ZINC predictor, a step by step tutorial: | |
| Step 1: Input/upload sequences for prediction | |
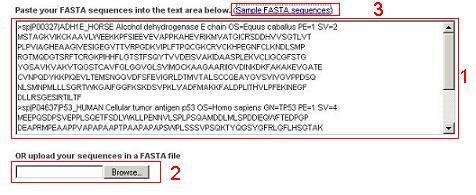 |
The predictor accepts protein sequences in FASTA format.
|
| Step 2: Select or input threshold parameters | |
 |
We provide three optimized settings for typical scenarios and users can also provide a custom threshold.
|
| Step 3: Choose output parameters | |
 |
We combined the predicted zinc-binding motif from Hufzinc predictor with a curated list of PFAM zinc finger HMMs. We provide four settings for displaying different results.
|
| Step 4: Reading the prediction result | |
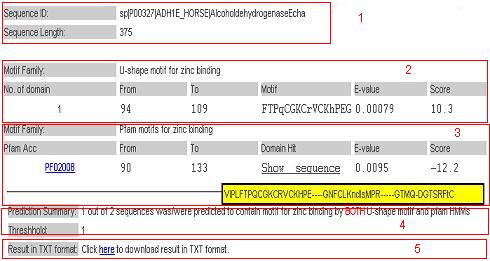 |
For each query sequence, we provide the following information according to the output option selected.
|
| Frequently Asked Questions: | ||
| Are there any limitations on the number of sequences/residues? | ||
| I have a very large dataset to be analyzed, how should I proceed? | ||
| Can I download the complete results in a tab-separated txt-format (for import in Excel or easier parsing)? | ||
| How to interpret the score of a predicted motif? | ||
| What are the PFAM zinc finger domains used in the website? | ||
| Can I download the positive and negative sequence sets used for training and the Huf-Zinc HMM? | ||
| Contact us | ||
| Return to Predictor | ||
| Are there any limitations on the number of sequences/residues? | ||
| There are no direct limitations on the number of sequences and residues. However, due to practical reasons, we have to limit the size of uploaded datafiles to 100MB, which nevertheless allows whole proteomes to be analyzed. | ||
| Return to Top Predictor | ||
| I have a very large dataset to be analyzed, how should I proceed? | ||
| If the dataset you would like to analyze exceeds the above mentioned 100MB filesize, please contact us directly for providing such predictions. | ||
| Return to Top Predictor | ||
| Can I download the complete results in a tab-separated txt-format (for import in Excel or easier parsing)? | ||
| Yes, at the very bottom of the output you can find a link to download the complete prediction results in a tab-separated txt-format. The format is one predicted motif per line and info in the following order: "query protein description line, motif desciption/pfam accession, motif start position, motif end postion, HMMER bit score, HMMER E-value, predicted motif". | ||
| Return to Top Predictor | ||
| How to interpret the score and E-value of a predicted motif? | ||
| The displayed prediction score is the HMMER bit score and the E-value is the HMMER E-value. As the U-shape zinc binding motif is rather short, E-value significance estimates by HMMER can be misleading. Therefore, we chose the score thresholds for the different prediction scenarios according to performance in a rigourous cross-validation benchmark. For more detail on HMMER, please check The HMMER User's Guide or The HMMER package website . | ||
| Return to Top Predictor | ||
| What are the PFAM zinc finger domains used in the website? | ||
| We searched the PFAM database for zinc fingers and selected 68 PFAM domains that contain a cysteine-motif known to bind zinc. Click here for the list of PFAM domains we selected. | ||
| Return to Top Predictor | ||
| Can I download the positive and negative sequence sets used for training and the Huf-Zinc HMM? | ||
| Yes, we make our datasets available for comparison and further research. Download our 191 positive ZN binding motifs here and the 247 negative sequence motifs here. We also make the resulting and used HMM available here | ||
| Return to Top Predictor | ||
| Contact us | ||
| For questions regarding the HUF-ZINC predictor, please contact Sebastian. | ||
 | ||
| Return to Top Predictor | ||