sebastianms@bii.a-star.edu.sg
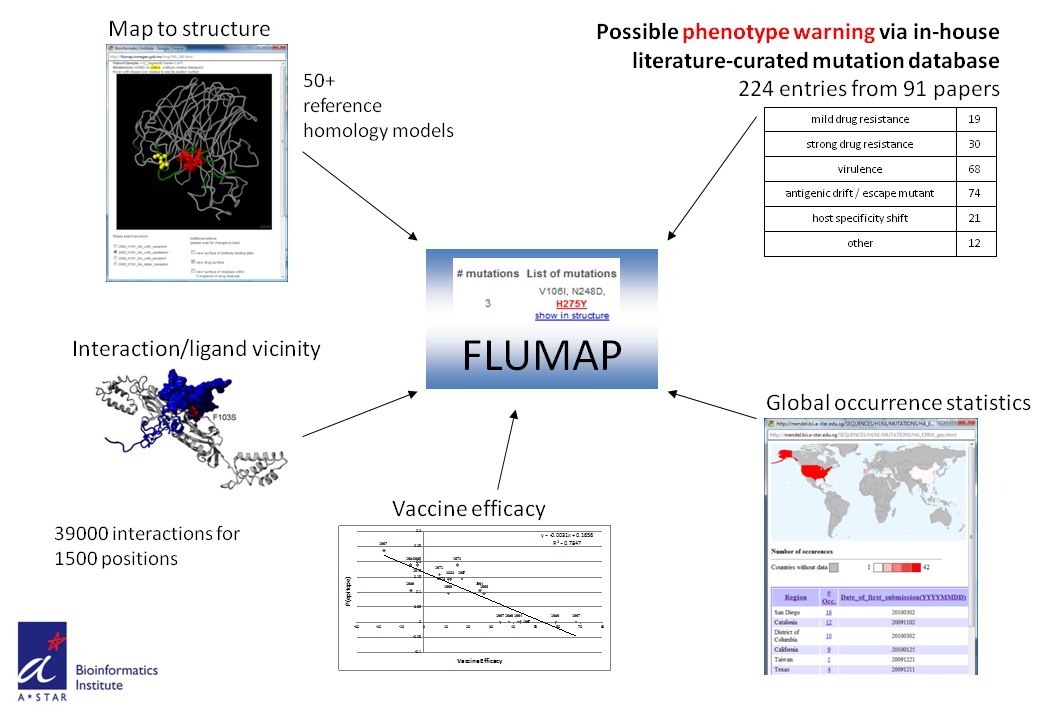
In short, a user can submit one or many nucleotide or protein sequences from influenza patient samples and the server will identify the best reference sequence to compare with and summarize all identified mutations. Each mutation is then annotated for their global occurrence and possible effect based on a set of literature-curated reports about equivalent mutations also from previous pandemics. The mutations are also presented in their structural context of known crystal structures or homology models, including the vicinity to ligands such as drugs, sialic acid or antibodies.
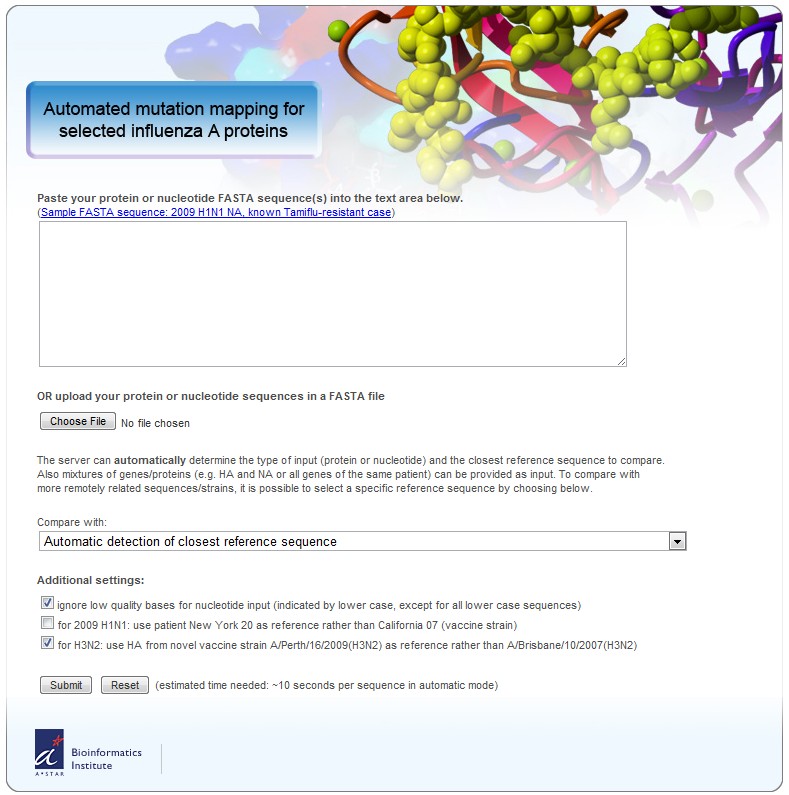
Usage is simple (2 steps), free for academic usage and the analysis quite exhaustive.
1. Submit your influenza sequence
2. Browse through the results providing detailed information for each mutation